More than 60 years after the discovery of the structure of the DNA molecule of life, it continues to surprise us and reveals its secrets one at a time. The double helix – elegantly simple and amazingly complex, now turns out to be far beyond just that!
From 1953 onward, everyone who has heard of DNA (the molecule that stores and transmits genetic code of almost all organisms on Earth) probably knows that it forms a helix of two complementary chains. The discovery made by the team of Dr. Watson and Dr. Crick remains one of the most important in modern science – they represent the model of so-called. B-DNA, which represents a right-rotating helix and is the form in which the DNA is located in the cells in normal conditions. And yet – since the confirmation of this model, several forms of double strands with different symmetry are observed in laboratory experiments – the most common are A- (more compact form of B-DNA) and Z- (left-rotating helix of DNA). While A-DNA is found in some extreme stressful conditions, the Z-DNA has always represented a serious problem for scientists – some believe that this form is accepted by DNA during transcription (one of the steps of protein synthesis), but the debate on the issue is yet continuing. Either way, the existence of DNA as a double helix is a fact accepted by both the scientific community and by the general public, but DNA continues to surprise us today with its diversity.
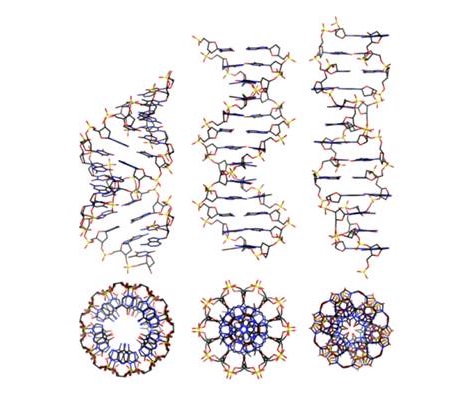
It turns out that DNA in living organisms can exist in many different forms and some of them considerably more complicated than just a double helix. A team from the University of Umeå, in Sweden, just this month proved that functional DNA can be found in the form of an impressive quadruple helix (quadruplex DNA). As it happens, the regions of DNA rich in guanine (one of the four bases coding genetic information) can form a large number of inter- and intramolecular quadruple spirals without violating the integrity of the DNA strands from which they are composed. Named guanine quartets (G4), those structures have been significantly enriched in specific regions of the genomes of many organisms (from yeast to human). They are rather more abundant in the region of the telomeres (the ends of chromosomes), the ribosomal DNA (part of DNA encoding RNA building up the ribosomes), and promoter regions (the areas acting as a power button for turning on/off the genes encoding the proteins).
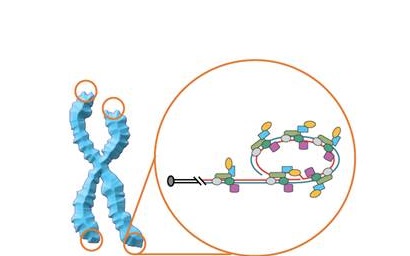
Telomeres – the linear strands represent DNA and the colorful shapes – different proteins binding to DNA in this region of the chromosome (Pic from http://goo.gl/6teHFU)
Scientists decided to explore the G4 stretches in yeast – Saccharomyces pombe. Their chromosomes are organized in a manner very similar to the human ones, and they are a widely used model organism for cellular, genetic and molecular experiments – a lot of structural and functional information for these organisms is already known, which makes exploring new phenomena a well controlled experiment.
The scientists from Umeå demonstrated that the G4 sequences, according to computer models, should form quadruple spirals, and later or showed that experimentally too. But the surprises do not stop there – they show that these sequences of DNA have a specific interaction with certain enzymes that operate the DNA in the cell. One of the important enzymes that manipulate DNA in the cell is the so called DNA helicase. Its function is to separate the two strands in the double helix during the replication of genetic information (DNA duplication for propagation of the cell or for the synthesis of proteins). Pfh1 helicase is used by the yeast, and it has its analogue in humans. It is responsible for unwinding DNA in areas where there are “obstacles” – proteins tightly bound to the DNA or compact DNA structures that require special attention, for example at the end of chromosomes (the so called telomeres regions). Pfh1 recognizes, binds to and is able to unravel G4 sequences in the DNA, the latter experiments have shown that this type of helicase is substantially concentrated in the regions of the telomeres during the S-phase (the phase of the cell cycle prior to cell division, when DNA is copied in order to provide both new cells with equal quantity and quality DNA). This unraveling of DNA G4 regions, however, is significantly slower (compared to unwinding double stranded DNA), which is probably a protective mechanism as is the very presence of G4 sequence in the telomeres.
Today we know that errors in the copying of telomere sequences or their significant shortening is among the most common factors in the development of tumors in multi-cellular organisms. Following this logic, cells have evolved sophisticated “security systems” against damage to the telomeres. Scientists suggest that structures such as quadruple spirals of G4 sequences may be one such a mechanism – if the ends of the chromosomal DNA are difficult to unwind, the likelihood for introducing random mutations or molecular breakage there is significantly reduced, and therefore the cell has a better chance of survival. On the other hand, the cell must still be capable in certain situations to operate with this DNA. To this end, the organisms evolved a special helicase, which handles puzzle-quadruple-spirals (Pfh1). Any complication of genetic machinery is expensive for the cell – it requires more energy and the emergence of a mutation in any of the genes coding for components of this machinery may be fatal, but it is a price that many organisms are ready to pay in order to protect the ends of their chromosomes.
Scientists were able to unravel one more of the many secrets of DNA and learned more about the cunning of living organisms in regard to protecting their genetic material from damage. The original article about quadruple helix and the ability of Pfh1 to deal with it came out in May 2016 – http://goo.gl/utXJZ9; and more information about Pfh1 helicase can be found here – http://goo.gl/n2ODiC.
