Evolution is not the means to an end
We often think wrongly about evolution as a process which is directional – headed in a specific “predetermined” way, the road to a specific endgame of Nature. But it is not. Evolution is the combination of naturally occurring variation, spontaneously or as a result of environmental pressure and extreme factors of the environment. It might seem as a witty process that “knows where it’s supposed to go” to us, just because we can observe the current snapshot in time of life, which is a product of millions of years of evolution. Evolution is not Nature’s toy-train that was meant to begin at the most simple cells roughly 4 billion years ago and end with its ultimate, most-complex creation – the human. Evolution is more of a kaleidoscope, which turns every time the conditions of the environment require so (a trait is selected for/against) and the picture changes – sometimes slightly, sometimes dramatically. There’s no final picture that will occur even if you turn the glass-lined tube a million times.
Another mistake we often make is, that we think of evolving organisms as ever-increasing in complexity. Some woos even talk of devolution, when the “next step” (the more recent organism) is more simple than the previous. Truth is, complexity is very biologically expensive to maintain, and if certain trait is for long time not required by the environment for survival, it might very well quickly be left behind in history, and the modern forms will evolve to utilize the energy they acquire for something different, more useful at the time.
Are humans smarter than Nature when in comes to evolution?
Directed evolution is a growing research field. Scientists use different methods to predict and streamline the evolution of DNA and proteins in order to improve traits or study evolutionary mechanisms. Scientists have been using it to create mostly enzymatic properties and substrate specifics, which did not exist naturally before. Some enzymes have been optimized to work more than 4000 times better than the original counterparts. After making anything work 4000 times better than before – the sky is the limit – quite literally! Other enzymes have been tweaked to perform their action without their usual side-reactions which lead to low yield and bring out the need for chemical purification of the main product of interest. This could make enzymatic production of compounds of interest much more cost, energy and time-effective process.
In fact, humans have been performing directed evolution experiments on a large-scale for millennia – breeding of plants and animals by farmers has been the ultimate field trial. By crossing individuals with preferable traits, farmers have been recreating at higher speed the natural process of creating genetic variation “at random”. Mixing whole genomes during the breeding programs much like during evolution – the effects are unpredictable. That is why after the crossing, comes the long sorting and selection of the most desired individuals. The DNA/proteins and respectively cells and organisms which are seen best – having more mass, or producing more milk, eggs, fruits etc, are taken to the next round of breeding and enhancing of traits.
We’ve been pushing the boundaries of Natural evolution ever since we got off the branch and into more settled lifestyle. But this method has its own limitations – only so much can one optimize an enzymatic active site – even the natural variation is not an inexhaustible tool. So we pushed a little further…
We didn’t only push the boundaries of evolution, we turned it inside out and still keep pushing
There are two general types of molecular symmetry when it comes to amino acids – D and L. Think of it as your left and right hands – they are exactly the same, but you cannon overlap them perfectly – there will always be one property that it will mismatch if you align all the rest.
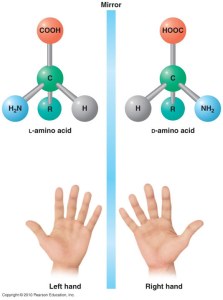
At the dawn of life, we are still not really sure how or why, the primitive life forms have “decided” to use the L-amino acids to build up their proteins. No protein is known to contain D-amino acids. In few rare occasions they have been found as parts of other biopolymers (such as the microbial cell wall sugars or some very short microbial peptides), but they stand more as the exception to prove the rule.
Prof Steven Kent and his colleagues went completely the other way compared to evolution. They started chemically synthesizing proteins entirely from D-amino acids. Now imagine the diversity of proteins throughout the living organisms and double this number! If you build a protein with exactly the same amino acid sequence, but use only D-amino acids, you’ll get the mirror image of your original protein. And from there on – the novel functionalities are truly novel.
Why do we need “out of this world” proteins?
Well to start with, the “we do it because we want to see if we can” of scientists have always pushed scientific and technological progress further, so scientific curiosity is a force to be reckoned with.
But there is more practical reason as to why would we attempt to restart the millions of years of evolution. Having the D-form next to the original L-form of proteins makes them much more easy to crystallize due to the perfect symmetry they offer. Crystallizing proteins is one of the most robust and widely used methods to study protein structure and deduct their functionality. Knowing the atomic structure of the active centers of enzymes or the binding pockets of antibodies is the first and most important step to optimizing their productivity and creating better biologicals for medical and industrial purposes.
And to up these stakes, if you think it’s not enough just yet, D-amino acid polypeptide chains can overcome the major problem of resistance. In all existing organisms, proteins have evolved to interact the way they do, based on the fundamental premise that they recognize each others’ fine differences in the 3D space. An enzyme recognizes one or few structurally similar substrates based on the fact that they fit in its active site. Receptors trigger cellular signals only when they are bound to their own specific epitope of another molecule. The ability of bacteria to evolve proteins which recognize and degrade other proteins is the basis of microbial immunity. Often, bacteria need to come up with an enzyme with only a few amino acid substitutions in order to be able to overcome a biological threat from an antibiotic – to evolve an iteration of the original form. This is one of the reasons why bacteria become so disturbingly quickly resistant to our latest generations of drugs. If all of a sudden we are able to produce anti-microbial peptide that these microbes have never even remotely encountered, it will take them much longer time to overcome this threat. Life always finds a way, so eventually they probably will find a neat little way to surprise us, but the time we’ll gain to keep advancing our ways in overcoming infection has no price!
So next time you think of evolution, think of it as the never-ending journey, and don’t be mistaken – we to can be very (r)evolutionary – if a scientist can imagine something, they’ll also find a way to do it!

Comments